Stochastic Network Interaction Model (SNIM) Tutorial
This is a beautifully simple ecological network model that can simulate predator-prey and competitive multispecies dynamics. This is the B model of the paper 1, I read it long ago and for many years I wanted to make a version of it, finally, I did it.
You can find the source code here https://github.com/lsaravia/snim. To install it, clone the repository using git clone or download and compile it, following the instructions in the README.
The model description from 1 is the following:
The second model again involves a set of S (possible) species plus empty sites (pseudospecies labelled 0). This set (the species pool) $\Omega(S)$ is then a discrete set:
$\Omega(S) = {0,1,2,…,S}$
where 0 indicates empty space, available to all species from the pool. Three basic types of transition are allowed to occur. Let us call $ A,B \in \Omega(S )$ two given spcies present at a given step. These possible transitions are summarized as follows.
(i) Immigration: an empty site is occupied by a species randomly chosen from the set of (non-empty) species, i.e. $A \in \Omega(S )-{0}$:
$0 \xrightarrow{u_i} A$
This occurs with a probability of colonization (of empty sites) $\mu_i$ . Notice that this colonization depends on the particular species.
(ii) Death: All occupied sites can become empty with some fixed probability $e_i$ :
$A \xrightarrow{e_i} 0$
(iii) Interaction: the same rule as in model A is used, but here the probability of success $P_{ij}$ is weighted by the coefficients of the interaction matrix. Here we use
$P_{ij} = \pi[\Omega_{ij} - \Omega_{ji}]$
where $\pi[x]=x$ when $x > 0$ and zero otherwise. This probability of an interaction occurring in the system between species $i$ and $j$ is a measure of the interaction strength linking interacting species.
Using SNIM from R
Once installed, you can easily run simulations from R.
First, the model requires two input files:
- A species parameter file (
.par) - A simulation parameter file (
.cfg)
Example: Species Parameter File
# Model parameters example
#
3 10000 # 3 species, total habitat 10000
0.1 0.1 0.1 # Immigration vector
1 1 1 # Extinction vector
# Omega interaction matriz
# Species 0 is empty space
# Species 1 Predator eats 2 and 3
# Species 2 has higher growth rate
# Species 3 outcompete species 2
0.0 0.0 0.0 0.0
0.0 0.0 3.0 2.0
4.0 0.0 0.0 0.0
2.0 0.0 0.5 0.0
Explanation:
- First line: Number of species and total habitat size.
- Second line: Immigration rates for each species.
- Third line: Extinction rates for each species.
- Following lines: Omega interaction matrix (including intrinsic growth rates and interactions).
Example: Simulation Parameter File
# Simulation parameters example
#
rndSeed = 0 # 0 means a random rndSeed
nEvals = 500 # number of time steps
tau = 0.01 # Integration step
iniCond = 1000 # Initial condition of abundance of species
# if only one number is specified all the species will have the same initial conditions
Running the Model from R
A set of R functions are included. First, set a global variable pointing to the SNIM binary. Then, run the simulation:
# Global path to the binary of SNIM model
#
snimBin <- "~/Dropbox/cpp/CaNew/snim/dist/Release/GNU-Linux/snim"
source("R/snim_functions.r")
out <- run_snim_plot("simul.cfg","model3.par","model.out")
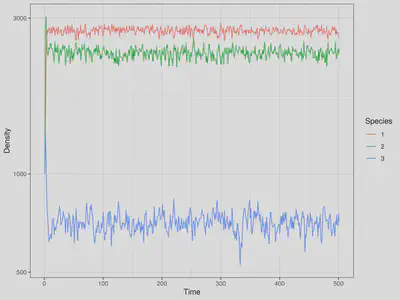
Animating the Dynamics with gganimate
An animation of the species densities can be created. First, set a shorter simulation (nEvals = 100), run the model again, and use:
require(gganimate)
ggplot(filter(out,Species!=0), aes(x = Time, y = Density,colour=Species)) +
geom_line() + theme_bw() + scale_y_log10() + xlab("Years") +
transition_reveal(Time)

Have fun!
-
Sole, R. V, Alonso, D. & Mckane, A. (2002). Self-organized instability in complex ecosystems. Philos. Trans. R. Soc. Lond. B. Biol. Sci., 357, 667–681 https://royalsocietypublishing.org/doi/10.1098/rstb.2001.0992. ↩︎ ↩︎